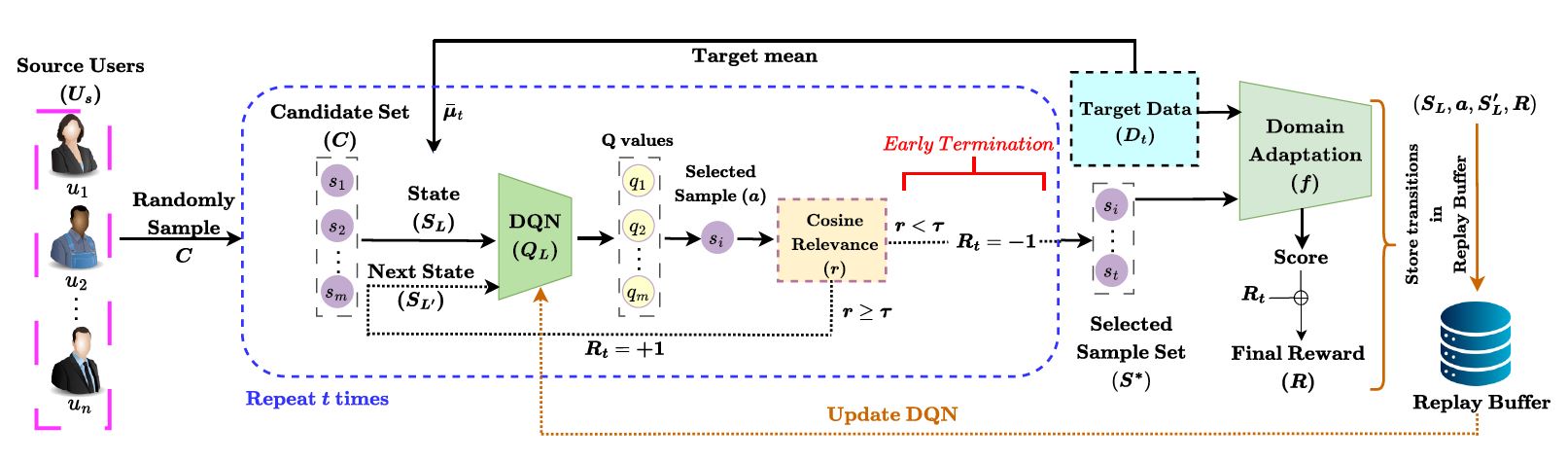
 | Accepted to ISBI 2026! Personalized Stress Detection Through Reinforcement Learning Based Domain Adaptation
Swati Kochhar, Nishchala Thakur, Katja Heilmann, Lejla Colic, Laith Hamid, Tara Chand, Martin Walter, Veronika Engert, and Deepti R. Bathula
ISBI 2026 abstract | bibtex Deep learning has achieved remarkable success in medical imaging; however, its reliance on large annotated datasets limits its applicability in data-scarce clinical settings. Few-shot learning addresses this limitation by enabling generalization from limited samples, yet conventional meta-learning approaches often overlook class-level heterogeneity and fail to exploit the rich metadata available in medical datasets. We propose Meta-data Class Sampling (MCS), a strategy that leverages dataset-level meta-information to guide class selection during episodic training. By identifying underperforming classes through per-class accuracy derived from metadata, MCS prioritizes challenging and semantically similar class combinations, thereby constructing more informative episodes. When integrated with Prototypical Networks, the proposed MCS module yields consistent performance gains, achieving average improvements of 8.10% on Derm7pt, 0.37% on ISIC-2019, and 3.69% on HAM10000.
@inproceedings{SK2026ISBI,
title={Personalized Stress Detection Through Reinforcement Learning Based Domain Adaptation},
author={Kochhar, Swati and Thakur, Nishchala and Heilmann, Katja and Colic, Lejla and Hamid, Laith and Tara Chand, Walter, Martin and Engert, Veronika and Bathula, Deepti R.},
booktitle={ISBI},
year={2026}} |
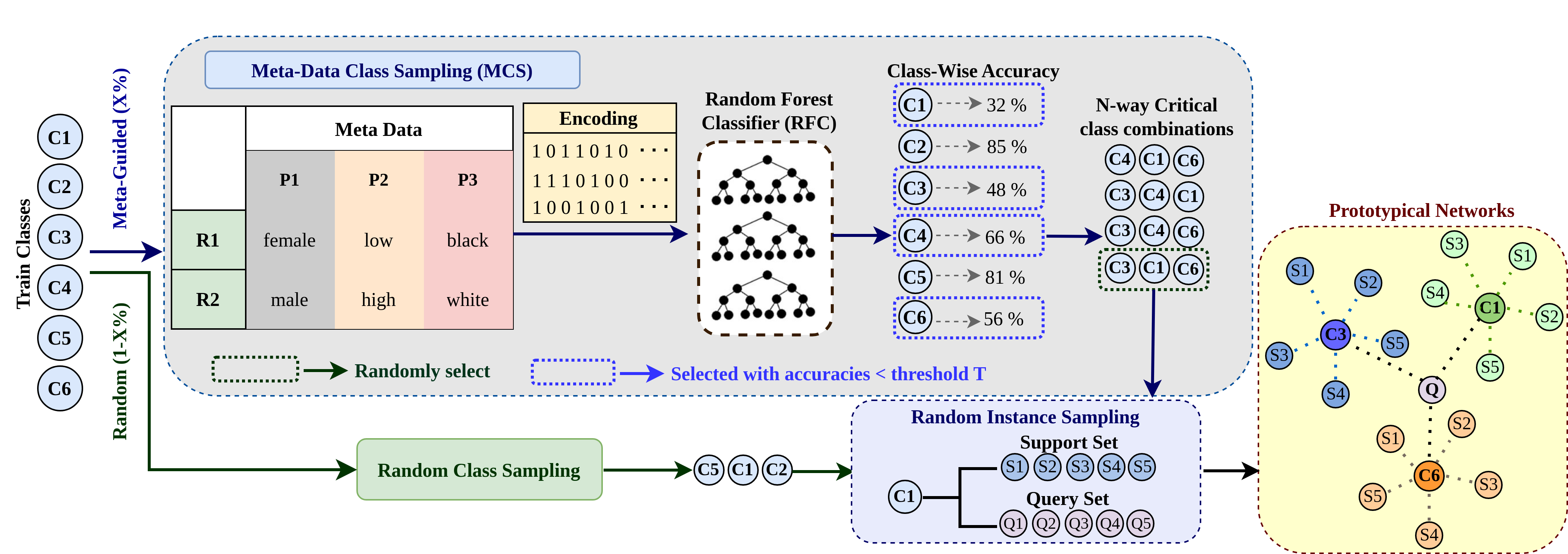 | Accepted to ISBI 2026! Meta-Guided Sampling for Improved Few-Shot Learning in Skin Cancer Classification
Ranjana Roy Chowdhury and Deepti R. Bathula
ISBI 2026 |
 | Congratulations to Usma Niyaz! Awarded Society of Women Engineers (SWE) Scholarship 2025 - 2026 |
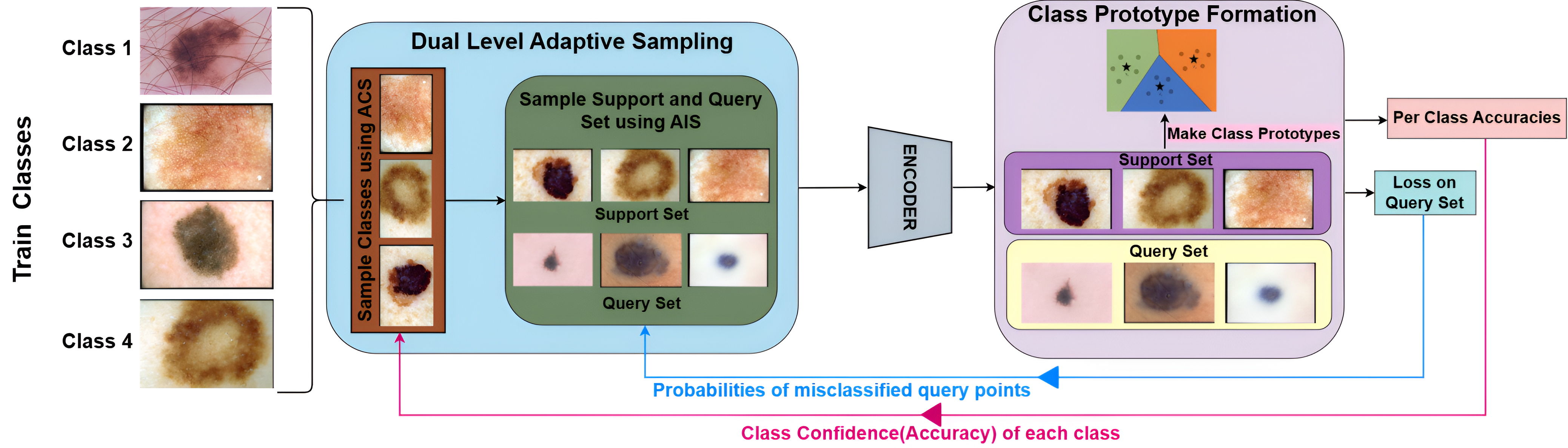 | Best Student Paper Award! Dual-Level Adaptive Sampling for Enhanced Few-Shot Medical Image Classification
Ranjana Roy Chowdhury, Usma Niyaz and Deepti R. Bathula
7th IEEE Symposium on Computers & Informatics (ISCI) 2025 abstract | bibtex Medical image classification is often hindered by limited labeled data, making few-shot learning (FSL) a crucial approach. However, conventional FSL methods struggle with the high variance present in medical images. Consequently, we propose Dual-Level Adaptive Sampling (DLAS) to enhance FSL by strategically selecting the most informative class combinations and instances within each class. During training, our class sampling policy prioritizes challenging, underconfident classes to foster continual learning. Subsequently, instances within each class are weighted according to their true class probability, ensuring the selection of hard-to-classify samples for improved generalization. This dual-level strategy is seamlessly integrated into prototypical networks within a metric-based meta-learning framework and can be adapted to various few-shot learning models. We validate our approach through extensive quantitative and qualitative analyses on three benchmark medical imaging datasets—Derm7pt, PathMNIST, and ChestMNIST. Results show that our sampling framework significantly improves model gen- eralization and classification performance compared to existing State-of-the-art methods.
@inproceedings{Chowdhury2025ISCI,
title={>Dual-Level Adaptive Sampling for Enhanced Few-Shot Medical Image Classification},
author={Chowdhury, Ranjana Roy and Niyaz, Usma and Bathula, Deepti R.},
booktitle={ISCI},
year={2025}} |
 | Congratulations to Abhishek Singh Sambyal! Awarded Science and Engineering Research Board (SERB) Internation Travel Grant to attend MICCAI 2024, Morocco. |
 | Congratulations to Abhishek Singh Sambyal! Awarded Medical Image Computing and Computer Assisted Intervention (MICCAI) 2024 RISE Registration Grant |
 | Congratulations to Ranjana Roy Chowdhury! Awarded Society of Women Engineers (SWE) Scholarship 2024 - 2025 |
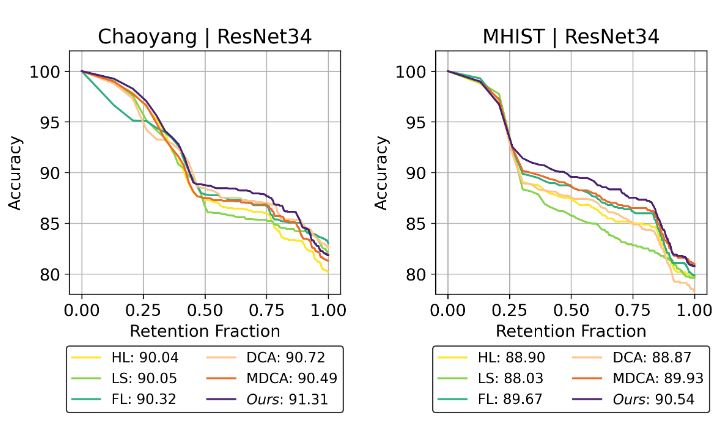 | Accepted to MICCAI 2024! LS+: Informed Label Smoothing for Improving Calibration in Medical Image Classification
Abhishek Singh Sambyal, Usma Niyaz, Narayanan C. Krishnan and Deepti R. Bathula
MICCAI 2024 abstract | bibtex Deep Neural Networks (DNNs) exhibit exceptional performance in various tasks; however, their susceptibility to miscalibration poses challenges in healthcare applications, impacting reliability and trustworthiness. Label smoothing, which prefers soft targets based on uniform distribution over labels, is a widely used strategy to improvemodel calibration. We propose an improved strategy, Label Smoothing Plus (LS+), which uses class-specific prior that is estimated from val- idation set to account for current model calibration level. We evaluate the effectiveness of our approach by comparing it with state-of-the-art methods on three benchmark medical imaging datasets, using two different architectures and several performance and calibration metrics for the classification task. Experimental results show notable reduction in calibration error metrics with nominal improvement in performance compared to other approaches, suggesting that our proposed method provides more reliable prediction probabilities. Our code will be made available on GitHub after acceptance.
@inproceedings{Sambyal2024MICCAI,
title={LS+: Informed Label Smoothing for Improving Calibration in Medical Image Classification},
author={Sambyal, Abhishek Singh and Niyaz, Usma and Krishnan, Narayanan C. and Bathula, Deepti R.},
booktitle={MICCAI},
year={2024}} |
 | Congratulations to Usma Niyaz! Awarded Science and Engineering Research Board (SERB) Travel Grant to attend ISBI 2024, Athens, Greece |
 | Congratulations to Usma Niyaz! Awarded 2024 IEEE International Symposium on Biomedical Imaging (ISBI) Travel Grant |
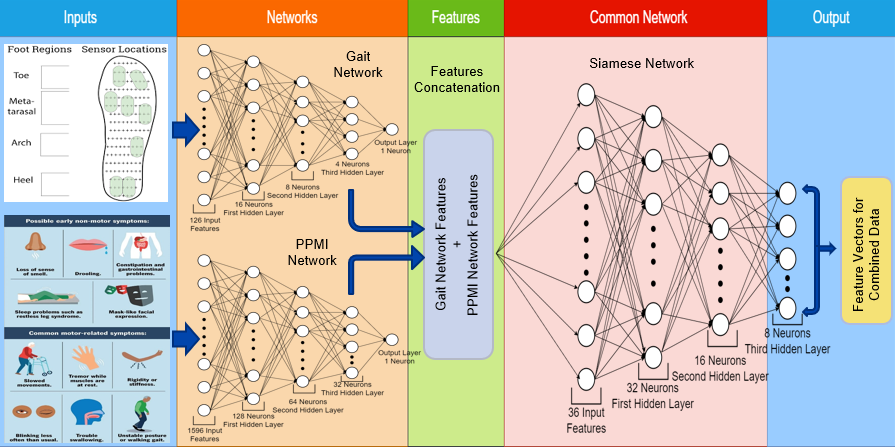 | Best Student Paper Award! Mutually Exclusive Multi-Modal Approach for Parkinson’s Disease Classification
Arunava Chaudhuri, Abhishek Singh Sambyal and Deepti R. Bathula
Bioimaging 2024 abstract | bibtex Parkinson's Disease (PD) is a progressive neurodegenerative disorder that affects the central nervous system and causes both motor and non-motor symptoms. While movement related symptoms are the most noticeable early signs, others like loss of smell can occur quite early and are easy to miss. This suggests that multi-modal assessment has significant potential in early diagnosis of PD. Multi-modal analysis allows for synergistic fusion of complementary information for improved prediction accuracy. However, acquiring all modalities for all subjects is not only expensive but also impractical in some cases. This work attempts to address the missing modality problem where the data is mutually exclusive. Specifically, we propose to leverage two distinct and unpaired datasets to improve the classification accuracy of PD. We propose a two-stage strategy that combines individual modality classifiers to train a multi-modality classifier using Siamese network with Triplet Loss. Furthermore, we use a Max-Voting strategy applied to Mix-and-Match pairing of the unlabelled test sample of one modality with both positive and negative samples from the other modality for test-time inference. We conducted experiments using gait sensor data (PhysioNet) and clinical data (PPMI). Our experimental results demonstrate the efficacy of the proposed approach compared to the state-of-the-art methods using single modality analysis.
@inproceedings{Chaudhuri2024MEMMPD,
title={Mutually Exclusive Multi-Modal Approach for Parkinson’s Disease Classification},
author={Chaudhuri, Arunava and Sambyal, Abhishek Singh and Bathula, Deepti R.},
booktitle={Bioimaging},
year={2024}} |
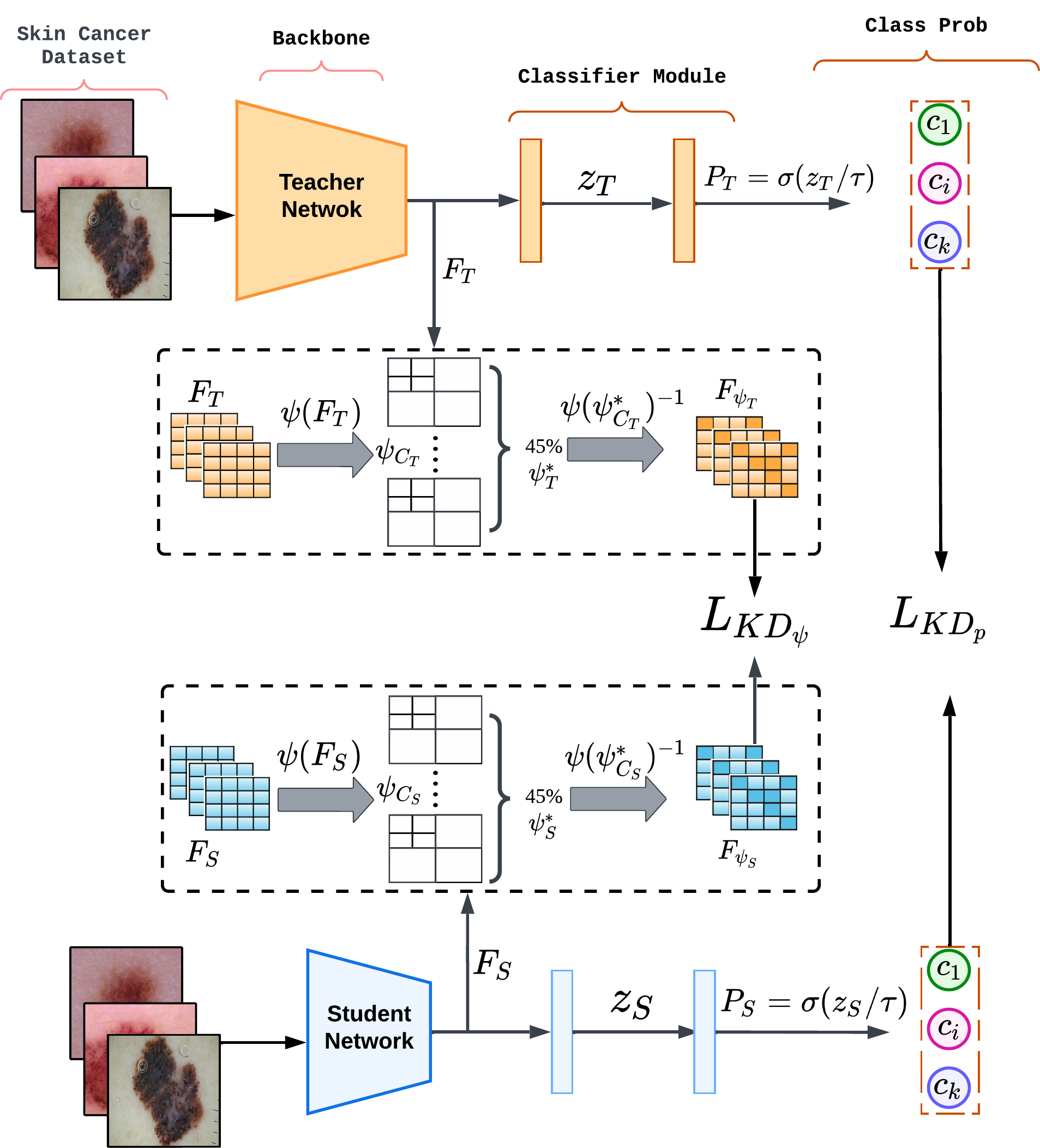 | Accepted to ISBI 2024! Wavelet-Based Feature Compression For Improved Knowledge Distillation
Usma Niyaz, Abhishek Singh Sambyal and Deepti R. Bathula
21st IEEE International Symposium on Biomedical Imaging 2024 abstract | bibtex Deep learning (DL) models can achieve state-of-the-art performance but at the cost of high computation and memory requirements. Due to their large capacity, DL models have the tendency to learn redundant features. In this work, we exploit this redundancy to improve model compression. Knowledge distillation (KD) aims to achieve model compression by transferring knowledge from a large, complex model to a simple, lightweight model. We propose an enhanced KD strategy that improves the efficiency of the distillation process by compressing the feature maps using Discrete Wavelet Transformation DWT, which helps capture crucial features from complex biomedical signals. Retaining and sharing only the most informative and discriminating features facilitates more effective feature mimicking. Extensive experiments using two benchmark datasets for Melanoma and Histopathology image classification tasks demonstrate the superiority of our proposed method over existing techniques. We further establish the generalizability and robustness of our method using two different teacher-student architectures and ablation studies.
@inproceedings{Niyaz2024ISBI,
title={Wavelet-Based Feature Compression For Improved Knowledge Distillation},
author={Niyaz, Usma and Sambyal, Abhishek Singh and Bathula, Deepti R.},
booktitle={ISBI},
year={2024}}
|